Here, we developed a comprehensive database of human transcriptional regulation of lncRNAs (TRlnc, http://www.licpathway.net/TRlnc), which aims to collect a large number of available resources of transcriptional regulatory regions of lncRNAs, and to annotate and illustrate their potential roles in the regulation of lncRNAs in cell type-specific manner.
The current version of TRlnc contains 8,683,028 typical enhancers/super-enhancers and 32,348,244 chromatin accessibility regions associated with 91,906 human lncRNAs. These regions were identified from over 900 human H3K27ac ChIP-seq, ATAC-seq and DNase-seq samples. Furthermore, TRlnc provides the detailed (epi) genetic information in transcriptional regulatory regions (promoter, enhancer/super-enhancer and chromatin accessibility regions) of lncRNAs, including common SNPs, Risk SNPs, eQTLs, Linkage disequilibrium SNPs, TFs predicted by motifs, TFs identified by ChIP-seq, methylation sites of 450K array, methylation sites of whole-genome shotgun bisulfite sequencing, histone modifications and 3D chromatin interactions, especially supporting the display of TFs binding to transcriptional regulatory regions from over 7,000 TF ChIP-seq samples. In addition, TRlnc integrates expression, diseases and miRNAs associated with lncRNAs from multiple sources.

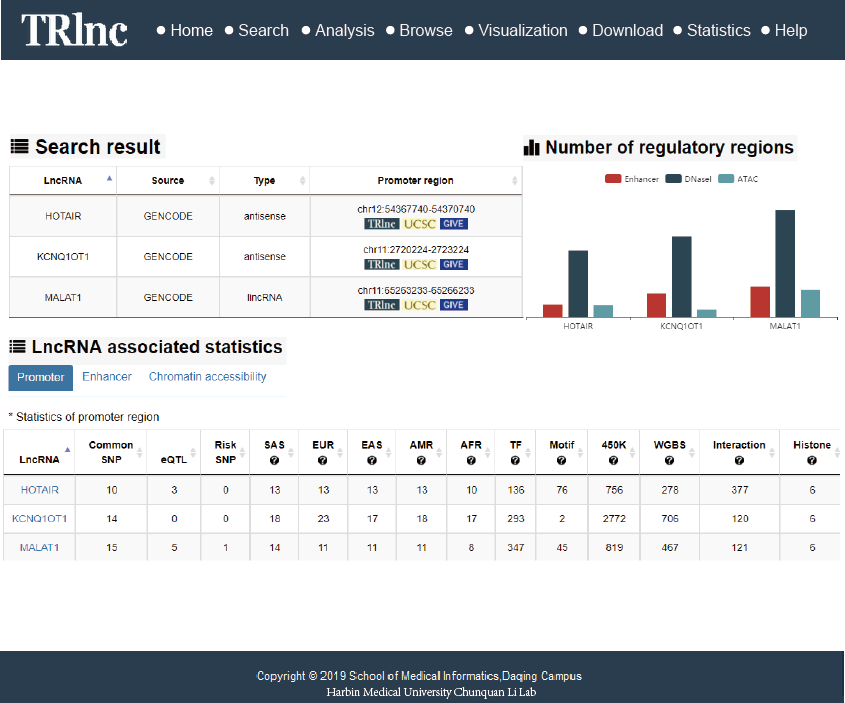
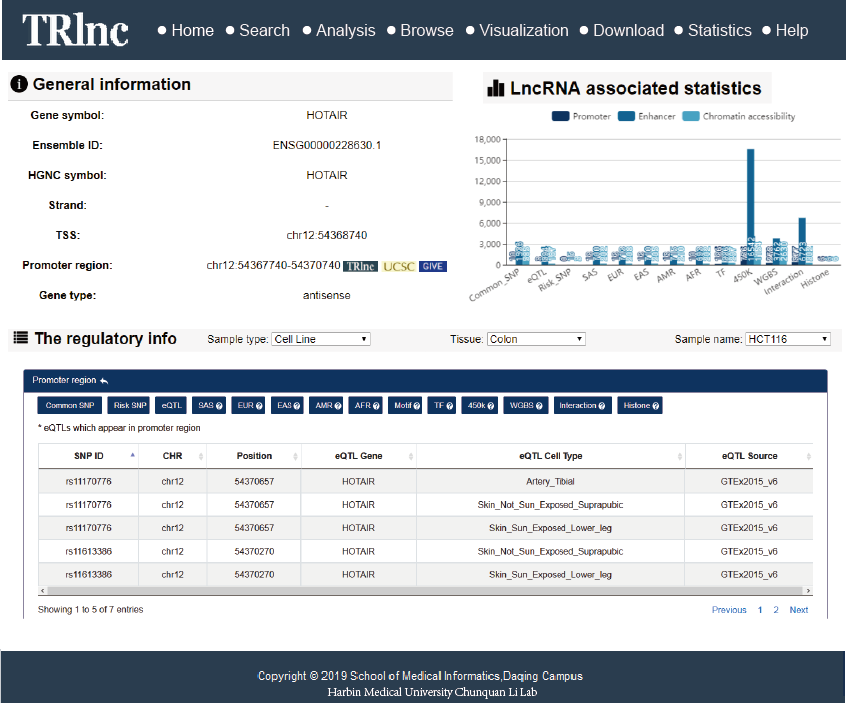
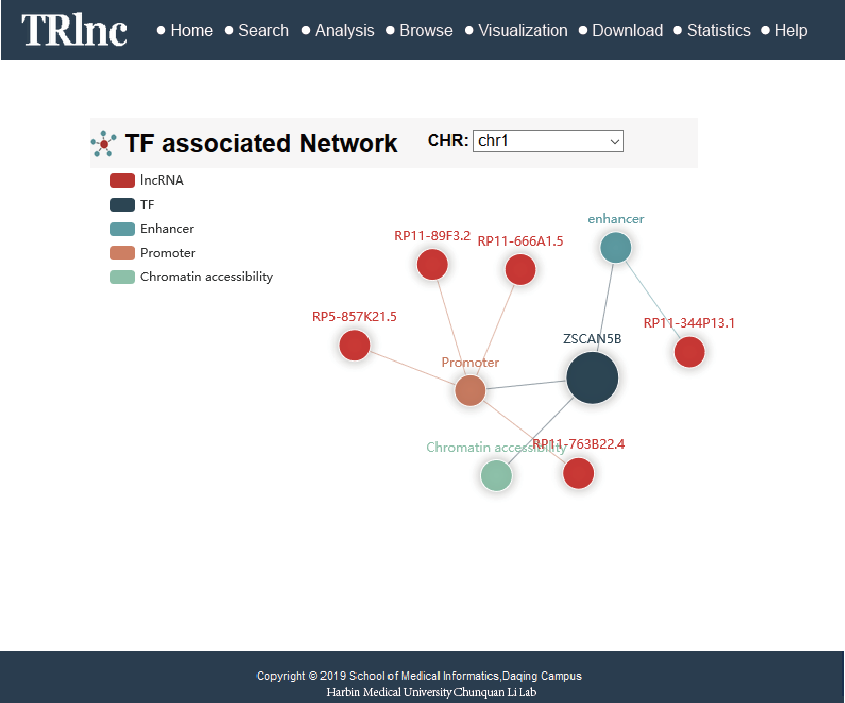
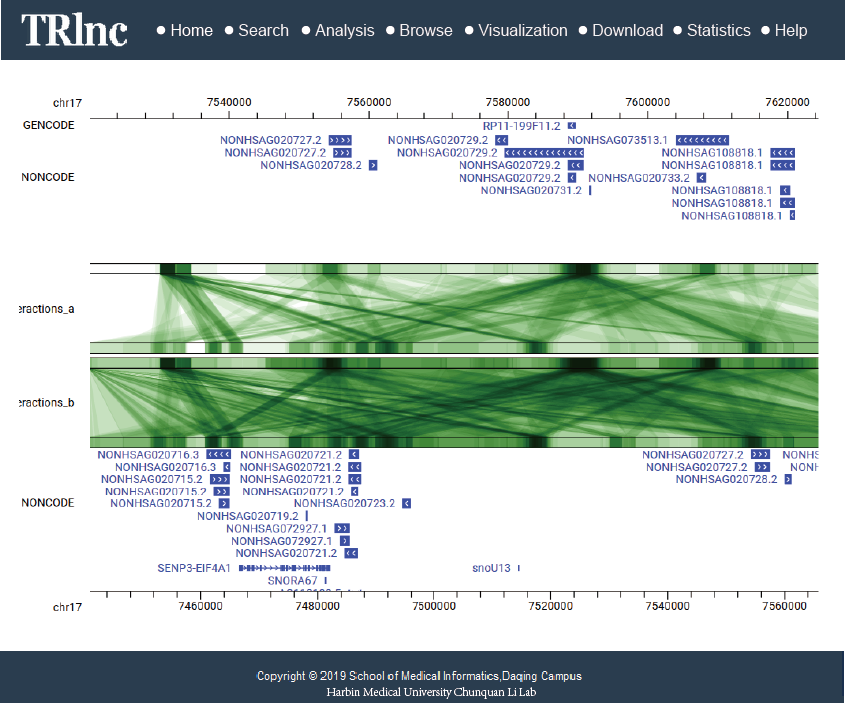
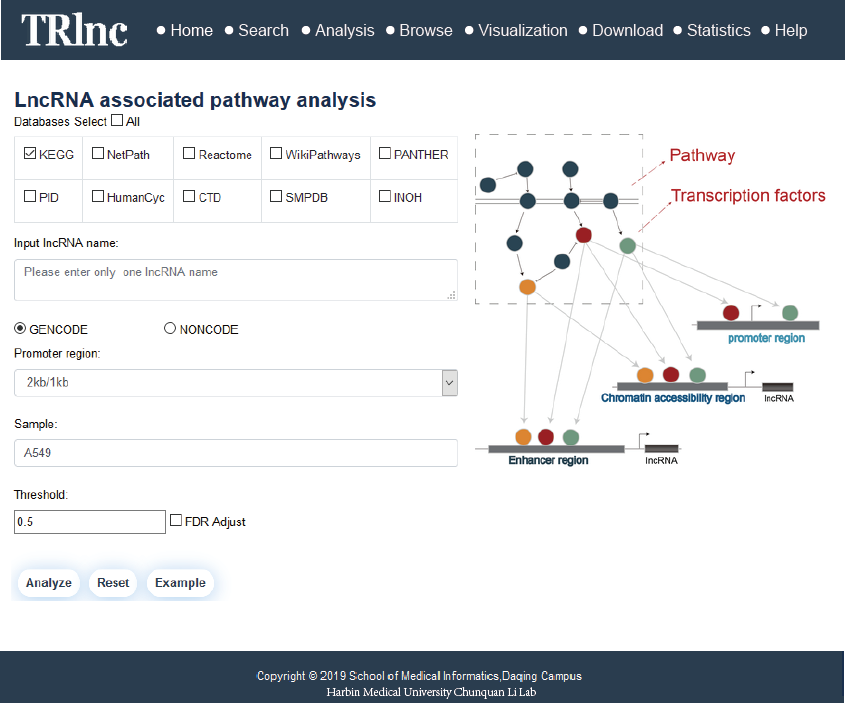
 Transcriptional regulatory regions of lncRNAs
Transcriptional regulatory regions of lncRNAs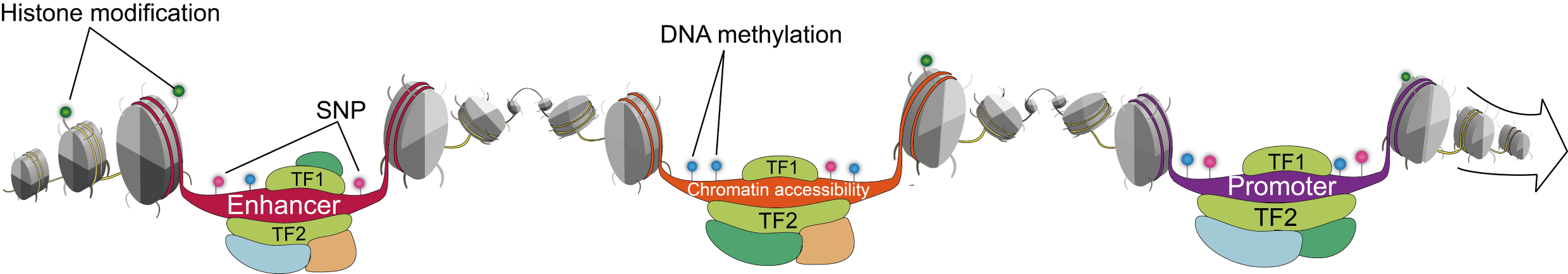
 Data statistical table
Data statistical table Contact us
Contact us Visitors
Visitors